|
|
|
Interferon was scarce and expensive until 1980, when the interferon gene was inserted into bacteria using recombinant DNA technology, allowing for mass cultivation and purification from bacterial cultures.
This year, an estimated 1.4 million Americans will have a new or recurrent heart attack.
As many as 20% of Americans have been infected by the fungus known as Histoplasmosis. While most people are asymptomatic or only have slight symptoms, infection can progress to a rapid and potentially fatal superinfection.
As the western states of America were settled, pioneers often had to drink rancid water from ponds and other sources. This often resulted in chronic diarrhea, causing many cases of dehydration and death that could have been avoided if clean water had been available.
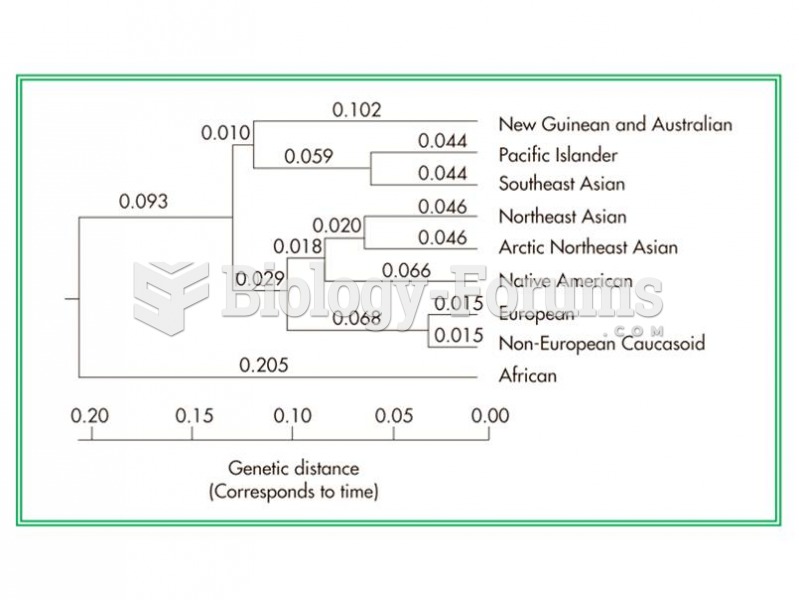
Stroke kills people from all ethnic backgrounds, but the people at highest risk for fatal strokes are: black men, black women, Asian men, white men, and white women.